This post explains how to work with a research compendium. The goal of a research compendium is to provide a standard and easily recognizable way for organizing the digital materials of a project to enable others to inspect, reproduce, and extend the research (Marwick B et al. 2018). A research compendium follows three general principles:
- Files are organized according to the conventions of the community
- Data, method, and output are clearly separated
- Computational environment that was used is specified
In other words, a research compendium is a simple way to organize files by separating the data, the code, and the results, while also documenting the computational environment.
This post is derived from the exercise proposed as part of the training course Reproducible Research in Computational Ecology.
Foreward
In order to assist us in creating the structure of our working directory, we will use the rcompendium package, developed by the author of this post. This package allows for the automation of creating files and directories specific to a research compendium (and a package).
The package is released on the CRAN but we will install the development version from GitHub:
If you encounter difficulties installing the package, please carefully read the Installation section of the README.
Once the package is installed, you need to run the set_credentials() function to store your personal information locally (first name, last name, email, ORCID, communication protocol with GitHub). This information will automatically populate certain files in the compendium. This function should only be used once.
This information has been copied to the clipboard. Paste its content into the file ~/.Rprofile (opened in RStudio using this function). This file is read every time is opened, and its content will be accessible to the rcompendium package.
Restart the session (Session > Restart R) and verify that your personal information is correctly accessible.
RStudio project
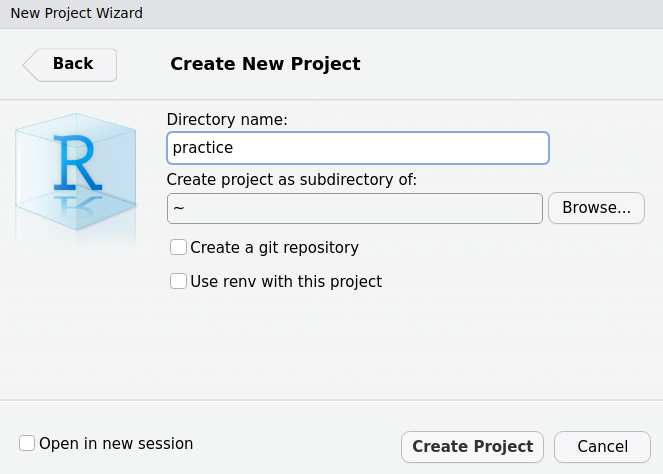
When you start a new project in it is strongly recommended to use RStudio Projects.
Create a new RStudio Project: File > New Project > New Directory > New Project and proceed as follow:
- choose a name for your project (short and without whitespace)
- select the location where the new project will be created
- uncheck all other boxes
- confirm
Always work within an RStudio project. This has the advantage of simplifying file paths, especially with the here package and its here() function. The paths will always be constructed relative to the folder containing the .Rproj file (the project root). This is called a relative path.
Never use the setwd() function again.
If you share your project on a cloud-based git repository (e.g. GitHub, GitLab, etc.) to collaborate, we recommend to add the .Rproj file to the .gitignore. The content of this .Rproj file can change between RStudio versions leading to unnecessary git conflicts. Listing the .Rproj file in the .gitignore will ensure that each user is working locally with its own version of this file.
Research compendium at this stage
practice/ # Root of the compendium
|
└─ practice.Rproj # RStudio project fileREADME
Every project must contain a README file. It is the showcase of the project. The roles of a README are multiple:
- Describe the project
- Explain its contents
- Explain how to install it
- Explain how to use it
It is a simple text file (plain text-based file) that can be written in plain text (README.txt), in simple Markdown (README.md), in R Markdown (README.Rmd), in Quarto (README.qmd), etc.
Here, you will create a README.md (simple Markdown) at the root of your project.
Use the utils::file.edit() function, which allows you to open a file in the RStudio editor. If the file doesn’t exist, it will also create it.
Run this line of code in the console: here::here("README.md") and try to understand what the here::here() function does.
Edit this README.md by adding the information that you find relevant.
Suggestion
# Practice
This project contains files to create a simple **research compendium** as
presented in the training course
[Reproducible Research in Computational Ecology](https://rdatatoolbox.github.io).
## Content
This project is structured as follow:
- `README.md`: presentation of the project
- `practice.Rproj`: RStudio project file
## Installation
Coming soon...
## Usage
Coming soon...
## Citation
> Doe J (2024) Minimal structure of a research compendium.Always add a README to help the user understand your project. If you want to execute code inside, write it in R Markdown (README.Rmd) or Quarto (README.qmd), otherwise, simply use basic Markdown (README.md).
NB. If you write a .Rmd or .qmd, don’t forget to convert it into a .md file. GitHub can only interpret basic Markdown.
You can also click on the Render button of RStudio.
rcompendium::add_readme_rmd() function
You can use the add_readme_rmd() function of rcompendium package that populates a README template for projects.
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
└─ README.md # Presentation of the projectDESCRIPTION
The DESCRIPTION file describes the metadata of the project (title, author, description, dependencies, etc.). It is one of the essential elements of a package. Here, we will repurpose it for use in a research compendium in order to take advantage of package development tools (see below).
Add a DESCRIPTION file using the add_description() function of the rcompendium package.
Package: practice
Type: Package
Title: The Title of the Project
Version: 0.0.0.9000
Authors@R: c(
person(given = "Jane",
family = "Doe",
role = c("aut", "cre", "cph"),
email = "jane.doe@mail.me",
comment = c(ORCID = "0000-0000-0000-0000")))
Description: A paragraph providing a full description of the project (on
several lines...)
License: {{license}}
Encoding: UTF-8As you can see, the DESCRIPTION file has been pre-filled with your personal information. You will edit the Title and Description fields later.
Always add a DESCRIPTION file at the root of the project. It is used to describe the project’s metadata: title, author(s), description, license, etc. We will discuss this later, but it is also the ideal place to list required external packages.
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
└─ DESCRIPTION # Project metadataLICENSE
Any material shared online must have a LICENSE that describes what can be done with it. Therefore, we recommend adding a license to your project from the start. To determine which license is most appropriate for your project, you can visit this website: https://choosealicense.com.
Add the GPL-3 license to your project using the add_license() function of the rcompendium package.
Note that a new file has been created: LICENSE.md. This file details the contents of the license and will be read by GitHub. Also, check the content of the DESCRIPTION file: the License section has been updated thanks to rcompendium.
Add a section in the README.md mentioning the license.
Suggestion
# Practice
This project contains files to create a simple **research compendium** as
presented in the training course
[Reproducible Research in Computational Ecology](https://rdatatoolbox.github.io).
## Content
This project is structured as follow:
- `README.md`: presentation of the project
- `DESCRIPTION`: project metadata
- `LICENSE.md`: license of the project
- `practice.Rproj`: RStudio project file
## Installation
Coming soon...
## Usage
Coming soon...
## License
This project is released under the
[GPL-3](https://choosealicense.com/licenses/gpl-3.0/) license.
## Citation
> Doe J (2024) Minimal structure of a research compendium.Always add a LICENSE to a project that will be made public. Visit the Choose a License website to select the most appropriate one for your project.
Note: If no license is provided, your project will be subject to the No License rules: no permissions are granted. In other words, no one can do anything with your project (no reuse, no modification, no sharing, etc.).
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
└─ LICENSE.md # License of the projectSubdirectories
The next step involves creating subdirectories, each with a specific role. The idea here is to separate the data, results, and code.
To do this, use the add_compendium() function from rcompendium.
A good Research compendium will consist of different subdirectories, each intended to hold a specific type of file. By default, the add_compendium() function will create this organization:
- The
data/folder will contain all the raw data necessary for the project. - The
outputs/folder will contain all the generated results (excluding figures). - The
figures/folder will contain all the figures produced by the analyses. - The
R/folder will only contain functions (and their documentation). See below for more details. - The
analyses/folder will contain scripts (or.Rmdand/or.qmdfiles) that will call the functions.
Note: This structure can of course be adapted based on needs, personal practices, and the complexity of the project. With the exception of the R/ folder, all other directories can be named differently.
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
├─ outputs/ # Contains results
├─ figures/ # Contains figures
├─ R/ # Contains R functions (only)
└─ analyses/ # Contains R scriptsWriting code
We’re ready to code!
Here we will write a code that will download the PanTHERIA dataset (Jones et al. 2009) and save it locally in our compendium.
PanTHERIA is a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Metadata can be found here. Note that missing values are coded -999.
We’ll start by writing our code in a script. The PanTHERIA data file, available here, will be saved in the data/pantheria/ subdirectory.
Create the download-data.R script in the analyses/ directory using the utils::file.edit() function.
Now write the code to download the data file.
Use dir.create() to create the subdirectory data/pantheria/, here::here() to build robust paths and utils::download.file() to download the file from the URL.
Suggestion
# Download PanTHERIA dataset
#
# Author: Jane Doe
# Date: 2024/09/24
## Destination path ----
path <- here::here("data", "pantheria")
## Create destination directory ----
dir.create(path, showWarnings = FALSE, recursive = TRUE)
## File name ----
filename <- "PanTHERIA_1-0_WR05_Aug2008.txt"
## Repo base URL ----
base_url <- "https://esapubs.org/archive/ecol/E090/184/"
## Build full URL ----
full_url <- paste0(base_url, filename)
## Build full path ----
dest_file <- file.path(path, filename)
## Download file ----
utils::download.file(url = full_url,
destfile = dest_file,
mode = "wb")Try scripting the whole project (including data acquisition). Here, we’ve seen how to create files (utils::file.edit()) and directories (dir.create()), build robust relative paths (here::here()) and download files (utils::download.file()) directly from .
To use a function from an external package, you’ve learned to use library(pkg). In , there’s another syntax for calling a function from an external package: pkg::fun(). Whereas library() loads and attaches a package (making its functions directly accessible with fun()), the syntax pkg::fun() only loads a package in the session, but does not attach its contents. This means you have to specify the package name when calling the function.
We recommend using the pkg::fun() syntax. There are two reasons for this:
- A better code readability: at a glance, you’ll know which package the function is in.
- Limits conflicts between packages: two functions can have the same name in two different packages. For example, the
dplyrpackage offers afilter()function which is also found in thestatspackage (attached to the opening of ). However, thefilter()functions in these two packages do not do the same thing.
If you use library(dplyr), you’ll never be 100% sure whether you’re using the filter() function of the dplyr package or that of the stats package.
However, for very verbose packages (such as ggplot2), you can use the library() function, otherwise your code will quickly become tedious to write.
If you wish to use the %>% pipe, attach the magrittr package with library(magrittr).
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
| └─ pantheria/ # PanTHERIA database
| └─ PanTHERIA_1-0_WR05_Aug2008.txt
|
├─ outputs/ # Contains results
├─ figures/ # Contains figures
├─ R/ # Contains R functions (only)
|
└─ analyses/ # Contains R scripts
└─ download-data.R # Script to download raw dataCode refactoring
We can take this a step further by converting the script into function: this is known as code refactoring. A function is a set of lines of code grouped together in a single block to perform a specific task. Writing functions will make your code clearer and more easily reusable between projects.
Always store your functions (and only functions) in a directory named R/ located at the root of the project.
Convert the previous code into a function named dl_pantheria_data().
Use the usethis::use_r() function to create the function file inside the R/ directory.
Suggestion
dl_pantheria_data <- function() {
## Destination path ----
path <- here::here("data", "pantheria")
## Create destination directory ----
dir.create(path, showWarnings = FALSE, recursive = TRUE)
## File name ----
filename <- "PanTHERIA_1-0_WR05_Aug2008.txt"
## Repo base URL ----
url <- "https://esapubs.org/archive/ecol/E090/184/"
## Build full URL ----
full_url <- paste0(base_url, filename)
## Build full path ----
dest_file <- file.path(path, filename)
## Download file ----
utils::download.file(url = full_url,
destfile = dest_file,
mode = "wb")
return(dest_file)
}Write functions: this is called code refactoring. This will make your code clearer and easier to reuse. Always store your functions in the R/ folder. If you’re using functions from external packages, write them as follows: pkg::fun().
Finally adapt the content of the analyses/download-data.R script created earlier so that it calls the dl_pantheria_data() function.
Suggestion
# Download project raw data
#
# This script will download the PanTHERIA dataset and will store it in `data/`
# by calling the dl_pantheria_data() function available in the `R/` directory.
#
# Author: Jane Doe
# Date: 2024/09/24
## Download PanTHERIA dataset ----
pantheria_path <- dl_pantheria_data()The analyses/ directory contains scripts that call functions stored in the R/ folder. In the case of complex analyses, don’t hesitate to multiply the scripts (rather than having a single large script).
%%{init:{'theme':'neutral','flowchart':{'htmlLabels':false}}}%%
flowchart LR
B("analyses/download-data.R")
B --> C("dl_pantheria_data()")
Research compendium at this stage
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
| └─ pantheria/ # PanTHERIA database
| └─ PanTHERIA_1-0_WR05_Aug2008.txt
|
├─ outputs/ # Contains results
├─ figures/ # Contains figures
|
├─ R/ # Contains R functions (only)
| └─ dl_pantheria_data.R # Function to download PanTHERIA data
|
└─ analyses/ # Contains R scripts
└─ download-data.R # Script to download raw dataDocumentation
It’s time to document your function. It’s essential! To do this, we’re going to use the roxygen2 syntax. This makes it easy to document functions by placing a special header before the function. This header must contain (as a minimum) a title, a description of each argument and the function’s return.
Add a roxygen2 header to your function to document it.
Suggestion
#' Download PanTHERIA dataset
#'
#' @description
#' This function downloads the PanTHERIA dataset (text file) available at
#' <https://esapubs.org/archive/ecol/E090/184/PanTHERIA_1-0_WR05_Aug2008.txt>.
#'
#' The file `PanTHERIA_1-0_WR05_Aug2008.txt` will be stored in
#' `data/pantheria/`. Note that this folder will be created if required.
#'
#' @return This function returns the path (`character`) to the downloaded file
#' (e.g. `data/pantheria/PanTHERIA_1-0_WR05_Aug2008.txt`).
dl_pantheria_data <- function() { ... } Our function does not contain any parameter. But if this were the case, we would have had to describe the parameters with the roxygen2 tag #' @param.
Think of others (and of your future self)! Always document your code. Code without documentation is useless. Use roxygen2 headers to document your functions, simple comments to document code and README for everything else.
You can convert your roxygen2 headers into .Rd files, the only files accepted by for documenting functions. These .Rd files will be stored in the man/ folder. This is not mandatory when working with a research compendium but this is required if you develop a package.
Help for your function will be available via ?fun_name.
Research compendium at this stage (same as before)
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
| └─ pantheria/ # PanTHERIA database
| └─ PanTHERIA_1-0_WR05_Aug2008.txt
|
├─ outputs/ # Contains results
├─ figures/ # Contains figures
|
├─ R/ # Contains R functions (only)
| └─ dl_pantheria_data.R # Function to download PanTHERIA data
|
└─ analyses/ # Contains R scripts
└─ download-data.R # Script to download raw dataDependencies
Our project depends on two external packages: utils and here. As mentioned previously, the DESCRIPTION file is the ideal place to centralize the list of required packages.
Add these two dependencies to the DESCRIPTION file with the usethis::use_package() function.
Look at the contents of the DESCRIPTION file: the two required packages are listed in the Imports section.
Package: practice
Type: Package
Title: The Title of the Project
Version: 0.0.0.9000
Authors@R: c(
person(given = "Jane",
family = "Doe",
role = c("aut", "cre", "cph"),
email = "jane.doe@mail.me",
comment = c(ORCID = "0000-0000-0000-0000")))
Description: A paragraph providing a full description of the project (on
several lines...)
License: GPL-3
Encoding: UTF-8
Imports:
here,
utilsAlways list the required packages in the DESCRIPTION file. In this way, you will centralize the list of required packages in one place and use the devtools::install_deps() and devtools::load_all() functions (see section Loading the project).
If in your code you want to attach your packages with library(), use the usethis::use_package() function as follows:
The package will be added to the Depends section of the DESCRIPTION file.
Loading the project
Now that our compendium contains a DESCRIPTION file with a list of packages, we can use the package development tools available in the package devtools to:
1) Install packages with the devtools::install_deps() function
This function reads the DESCRIPTION file to retrieve packages listed in the Depends and Imports sections and install them (only if they are not already installed). This function therefore replaces the install.packages() function.
By default, this function will also ask you to update packages (if a new version is available). If you wish to disable this feature, add the argument upgrade = "never".
2) Load packages with the devtools::load_all() function
This function will read the DESCRIPTION file to retrieve packages listed in the Depends and Imports sections. It will load the packages listed in the Imports section and load and attach the packages listed in the Depends section. This function therefore replaces the library() function.
Update your DESCRIPTION file regularly by:
- adding any new packages you use
- removing packages you no longer use
3) Load functions with the devtools::load_all() function
The devtools::load_all() function has a second advantage: it will load functions stored in the R/ folder and make them accessible in the session. It therefore replaces the source() function.
After each modification to a function, don’t forget to execute the devtools::load_all() function. You can use the keyboard shortcut Ctrl + Shift + L in RStudio.
Try these two functions.
With a DESCRIPTION file (listing the required packages) and a R/ folder, you can use:
devtools::install_deps()to install (and update) packages: don’t useinstall.packages()anymore.devtools::load_all()to 1) load (and attach) packages and 2) load your functions: no longer uselibrary()orsource()(to load your functions).
Main script
To automate our project, we’ll create a main script at the root of the project. By convention, we’ll call it make.R. It will have two objectives:
- set up the project by installing and loading packages and functions
- run the project by sourcing scripts sequentially.
The idea is that, once the project is finished, the user only executes this script: it’s the conductor of the project.
Use the utils::file.edit() function to create a script at the root of the project.
Add the two previous functions:
Finally, add a line to the make.R file that will execute the analyses/download-data.R script.
Use the source() and here::here() functions to do this.
Suggestion
# Project title
#
# Project description
# ...
#
# Author: Jane Doe
# Date: 2024/12/02
# Setup project ----
## Install packages ----
devtools::install_deps(upgrade = "never")
## Load packages & functions ----
devtools::load_all()
# Run project ----
## Download raw data ----
source(here::here("analyses", "download-data.R"))A make.R file placed at the root of the project makes it easy to set up the project (install and load the required packages and functions) and run the various analyses sequentially (by sourcing scripts which themselves call functions). This is the conductor of the project.
Note: Given the simplicity of this project, we could easily have placed the contents of the script (analyses/download-data.R) in this make.R. The structure of a compendium is not fixed, but we recommend that you use at least functions and a make.R.
%%{init:{'theme':'neutral','flowchart':{'htmlLabels':false}}}%%
flowchart LR
A("make.R") --> B("analyses/download-data.R")
B --> C("dl_pantheria_data()")
Research compendium at the end
practice/ # Root of the compendium
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
| └─ pantheria/ # PanTHERIA database
| └─ PanTHERIA_1-0_WR05_Aug2008.txt
|
├─ outputs/ # Contains results
├─ figures/ # Contains figures
|
├─ R/ # Contains R functions (only)
| └─ dl_pantheria_data.R # Function to download PanTHERIA data
|
├─ analyses/ # Contains R scripts
| └─ download-data.R # Script to download raw data
|
└─ make.R # Script to setup & run the projectDocumentation (again)
Don’t forget to finalize your project documentation.
Edit the Title and Description sections of the DESCRIPTION file.
Package: practice
Type: Package
Title: Download PanTHERIA database
Version: 0.0.0.9000
Authors@R: c(
person(given = "Jane",
family = "Doe",
role = c("aut", "cre", "cph"),
email = "jane.doe@mail.me",
comment = c(ORCID = "0000-0000-0000-0000")))
Description: This project aims to download the PanTHERIA databases. It is
structured as a research compendium to be reproducible.
This is the result of the Practice 1 of the training course Reproducible
Research in Computational Ecology available at:
<https://rdatatoolbox.github.io/chapters/ex-compendium.html>.
License: GPL-3
Encoding: UTF-8
Imports:
here,
utilsFinally edit the README:
# Practice
This project aims to download the [PanTHERIA](https://doi.org/10.1890/08-1494.1)
database (Jones _et al._, 2009). It is structured as a research compendium
to be reproducible.
**NB.** This is the result of the Practice 1 of the training course
[Reproducible Research in Computational Ecology](https://rdatatoolbox.github.io).
## Content
This project is structured as follow:
.
|
├─ practice.Rproj # RStudio project file
|
├─ README.md # Presentation of the project
├─ DESCRIPTION # Project metadata
├─ LICENSE.md # License of the project
|
├─ data/ # Contains raw data
| └─ pantheria/ # PanTHERIA database
| └─ PanTHERIA_1-0_WR05_Aug2008.txt
|
├─ outputs/ # Contains results
├─ figures/ # Contains figures
|
├─ R/ # Contains R functions (only)
| └─ dl_pantheria_data.R # Function to download PanTHERIA data
|
├─ analyses/ # Contains R scripts
| └─ download-data.R # Script to download raw data
|
└─ make.R # Script to setup & run the project
## Installation
Coming soon...
## Usage
Open the `practice.Rproj` file in RStudio and run `source("make.R")` to launch
analyses.
- All packages will be automatically installed and loaded
- Datasets will be saved in the `data/` directory
## License
This project is released under the
[GPL-3](https://choosealicense.com/licenses/gpl-3.0/) license.
## Citation
> Doe J (2024) Download PanTHERIA and WWF WildFinder databases.
## References
Jone KE, Bielby J, Cardillo M _et al._ (2009) PanTHERIA: A
species-level database of life history, ecology, and geography of extant and
recently extinct mammals. _Ecology_, 90, 2648.
DOI: [10.1890/08-1494.1](https://doi.org/10.1890/08-1494.1)Congratulations
Your project is now a functional and reproducible research compendium.
The final compendium can be found here.
rcompendium::new_compendium() function
All these steps can be performed with a single function: new_compendium() from rcompendium. Read the documentation carefully before using this function.
References
Jones KE, Bielby J, Cardillo M et al. (2009) PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology, 90, 2648. DOI: https://doi.org/10.1890/08-1494.1.
Marwick B, Boettiger C & Mullen L (2018) Packaging data analytical work reproducibly using R (and friends). PeerJ. DOI: https://doi.org/10.7287/peerj.preprints.3192v2.