The purpose of the package snakedist is to compute the
distance between each pair of locations by following the shape of a
curved line (e.g. a river).
Setup
# Custom ggplot2 theme ----
custom_theme <- function() {
theme_light() +
theme(plot.title = element_text(face = "bold", family = "serif", size = 18),
plot.caption = element_text(face = "italic", family = "serif"),
axis.title = element_blank(),
axis.text = element_text(family = "serif"))
}Datasets
snakedist provides two datasets:
-
adour_lambert93: a spatial line layer (geopackage) of the French river L’Adour. -
adour_sites_coords: a table (csv) containing spatial coordinates of 5 locations sampled along the Adour.
First, let’s import the two datasets:
# Import the spatial layer of Adour river ----
path_to_file <- system.file("extdata", "adour_lambert93.gpkg",
package = "snakedist")
adour_river <- sf::st_read(path_to_file, quiet = TRUE)
adour_river
#> Simple feature collection with 1 feature and 1 field
#> Geometry type: LINESTRING
#> Dimension: XY
#> Bounding box: xmin: 334324.8 ymin: 6207072 xmax: 480886.8 ymax: 6308530
#> Projected CRS: RGF93 v1 / Lambert-93
#> river_name geom
#> 1 L'Adour LINESTRING (480886.8 620722...
# Import sites data ----
path_to_file <- system.file("extdata", "adour_sites_coords.csv",
package = "snakedist")
adour_sites <- read.csv(path_to_file)
adour_sites
#> site longitude latitude
#> 1 S-01 470911.2 6219515
#> 2 S-02 464962.6 6242684
#> 3 S-03 464023.4 6266791
#> 4 S-04 445238.4 6291838
#> 5 S-05 418626.3 6306239The function distance_along() (main function of the
package) requires that both layers are spatial objects. So we need to
convert the data.frame adour_sites into an
sf object.
# Convert data.frame to sf object ----
adour_sites <- sf::st_as_sf(adour_sites, coords = c("longitude", "latitude"),
crs = "epsg:2154")
adour_sites
#> Simple feature collection with 5 features and 1 field
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: 418626.3 ymin: 6219515 xmax: 470911.2 ymax: 6306239
#> Projected CRS: RGF93 v1 / Lambert-93
#> site geometry
#> 1 S-01 POINT (470911.2 6219515)
#> 2 S-02 POINT (464962.6 6242684)
#> 3 S-03 POINT (464023.4 6266791)
#> 4 S-04 POINT (445238.4 6291838)
#> 5 S-05 POINT (418626.3 6306239)Let’s map the data.
# Visualize data ----
ggplot() +
geom_sf(data = adour_river, col = "steelblue") +
geom_sf(data = adour_sites, shape = 19, size = 2) +
geom_sf_label(data = adour_sites, aes(label = site), nudge_x = 5000) +
geom_text(aes(x = 334500, y = 6285000), label = "L'Adour", hjust = 0,
color = "steelblue", fontface = "bold", size = 6, family = "serif") +
labs(caption = "RGF93 / Lambert-93 Projection") +
custom_theme()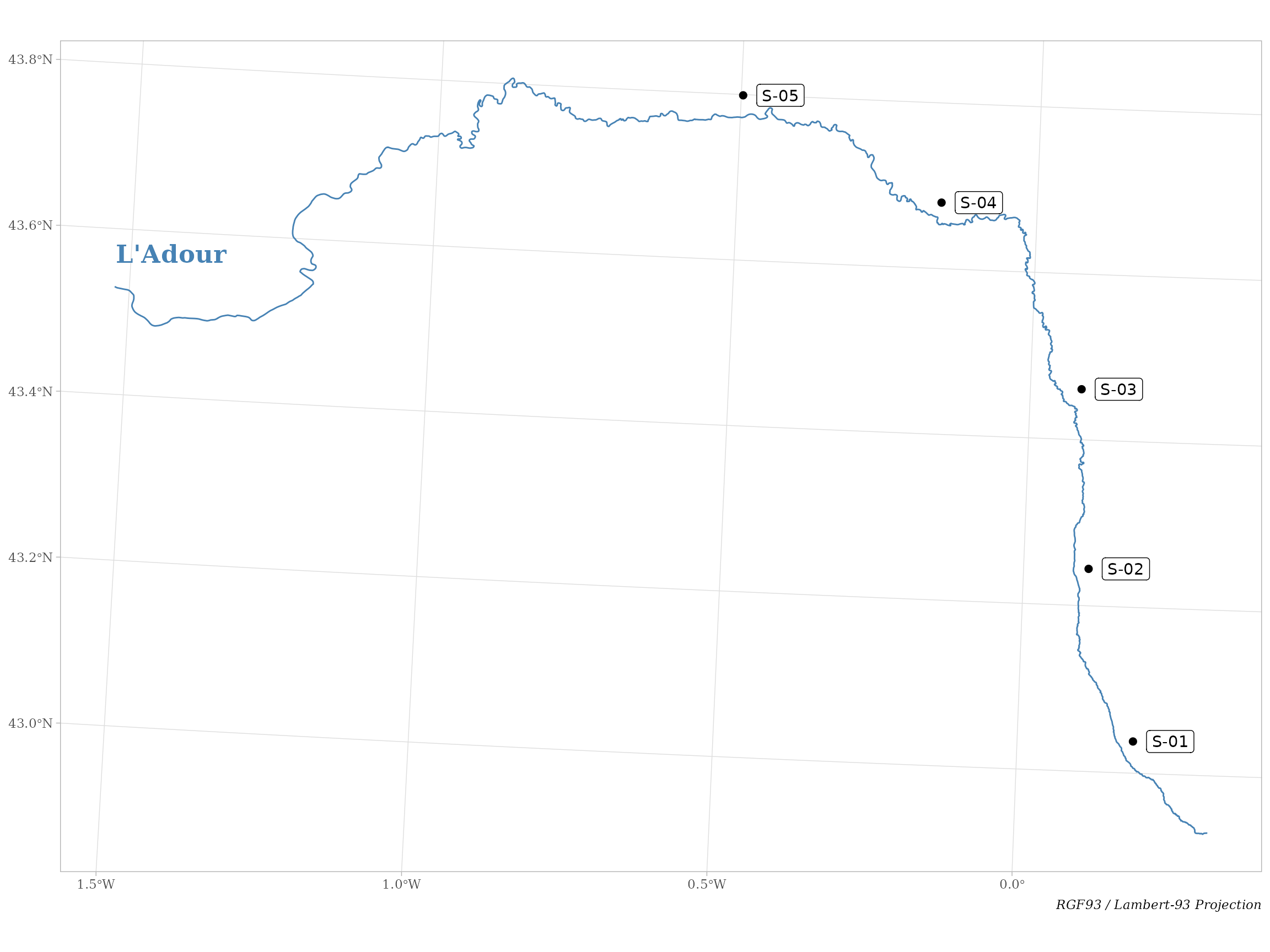
Figure 1. Study area with survey sampling
Compute distances
The function distance_along() computes the distance
between pairs of sites by following the shape of a linear structure (in
our case the river Adour). It uses the function
sf::st_line_sample() to regularly (or randomly, argument
type) sample points on the line with a given density of
points (argument density). All sampled points between the
two sites will be selected and their cumulative euclidean distance will
be computed.
# Compute distance along the river ----
dists <- distance_along(adour_sites, adour_river, density = 0.01, type = "regular")
head(dists, 12)#> from to distance
#> 1 S-01 S-01 0.00
#> 2 S-01 S-02 27422.59
#> 3 S-01 S-03 52913.17
#> 4 S-01 S-04 103064.90
#> 5 S-01 S-05 144881.86
#> 6 S-02 S-01 27422.59
#> 7 S-02 S-02 0.00
#> 8 S-02 S-03 25490.58
#> 9 S-02 S-04 75642.30
#> 10 S-02 S-05 117459.27
#> 11 S-03 S-01 52913.17
#> 12 S-03 S-02 25490.58This data.frame can be converted to a square matrix with
the function df_to_matrix().
# Convert to square matrix ----
df_to_matrix(dists)
#> S-01 S-02 S-03 S-04 S-05
#> S-01 0.00 27422.59 52913.17 103064.90 144881.86
#> S-02 27422.59 0.00 25490.58 75642.30 117459.27
#> S-03 52913.17 25490.58 0.00 50151.72 91968.69
#> S-04 103064.90 75642.30 50151.72 0.00 41816.97
#> S-05 144881.86 117459.27 91968.69 41816.97 0.00We can compare these values with the Euclidean distance.
# Compare to Euclidean distance ----
coords <- sf::st_coordinates(adour_sites)
dist(coords, upper = TRUE, diag = TRUE)
#> 1 2 3 4 5
#> 1 0.00 23919.63 47774.67 76743.68 101265.81
#> 2 23919.63 0.00 24125.69 52963.81 78653.68
#> 3 47774.67 24125.69 0.00 31308.31 60142.11
#> 4 76743.68 52963.81 31308.31 0.00 30259.12
#> 5 101265.81 78653.68 60142.11 30259.12 0.00This package is related to the package chessboard.