- Overview
- Features
- Installation
- First steps
- Long-form documentation (=vignettes)
- Citation
- Contributing
- Acknowledgments
- References
Overview
The package funbiogeo aims to help users with analyses in functional biogeography (Violle et al. 2014), the biogeography of species’ traits, by loading and combining data, exploring the relationships between traits and their availability trait coverage, providing many diagnostic plots to understand how to filter them, producing maps, correlating them with the environment, and helping to aggregate data at different scales It is aimed at first-timers of functional biogeography as well as more experienced users who want to obtain quick and easy exploratory plots.
Below is a quick introduction to the main features of funbiogeo, if you want some more details about them, check our vignettes.
Features
funbiogeo offers:
- Standardized functions to filter and select your data for further analyses,
- Pleasing default diagnostic plots to visualize the structure of your data,
- Extensive documentation (multiple vignettes, well-documented functions, real-life example dataset) to guide you through functional biogeography analyses,
- Nice default plotting functions fully compatible with the outputs of functional diversity packages (
betapart,fundiversity,hillR,mFD, etc.), - A publication-ready automated standardized report that provide analyses and plots of your data,
- Functions to easily “upscale” (=aggregate) your data to coarser spatial resolutions whatever the type of aggregation geometry you want (regular grid, irregular polygons, and rasters).

Naming scheme of available functions in funbiogeo
Installation
For the moment funbiogeo is not on CRAN but you can install the development version from R-universe as follow:
install.packages('funbiogeo', repos = c('https://frbcesab.r-universe.dev', 'https://cloud.r-project.org'))First steps
This section will show you some useful functions from funbiogeo. For a longer introduction please refer to the “Get started” vignette.
The package contains default example data named woodiv_traits, woodiv_site_species, and woodiv_locations all from the WOODIV database (Monnet et al. 2021). You can, for example, visualize the completeness of your trait dataset (which traits are known for which proportion of species) using the fb_plot_species_traits_completeness() function:
fb_plot_species_traits_completeness(woodiv_traits)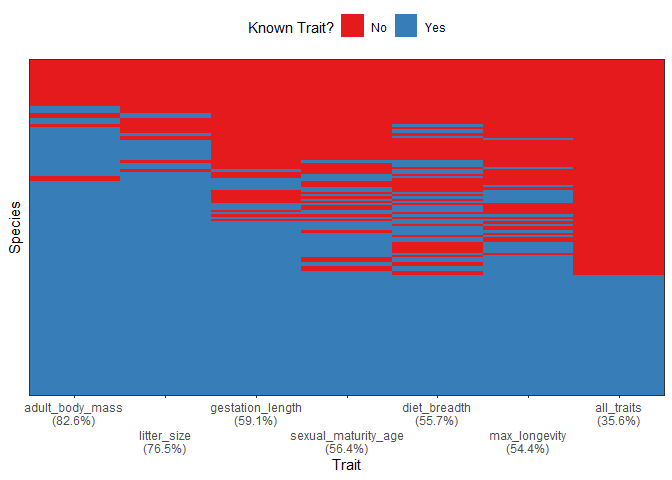
One other useful visualization is to see the trait coverage of each trait across all sites, using the function fb_map_site_traits_completeness():
fb_map_site_traits_completeness(woodiv_locations, woodiv_site_species, woodiv_traits)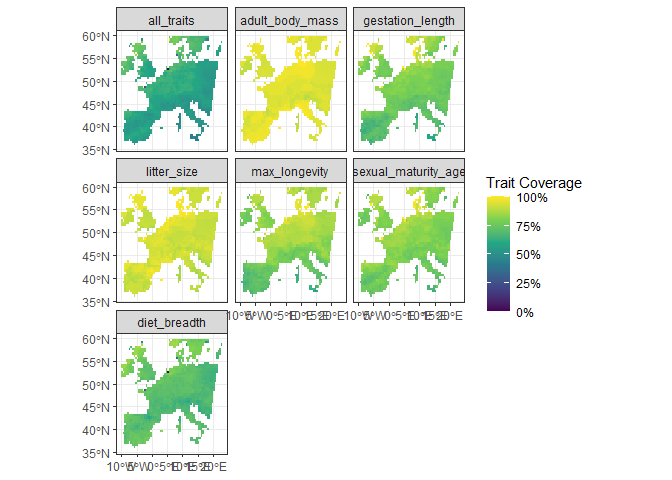
See more features of funbiogeo in the vignettes of the package
Long-form documentation
funbiogeo provides four vignettes to explain its functioning:
- A “Get started” vignette that describes its core features and guide you through a typical analysis.
- A vignette on all diagnostic plots provided in the package, which details how to use each plotting function and how to interpret their output.
- A vignette on the data format that
funbiogeoneeds, which shows you the use of specific functions to format your data to work well withinfunbiogeo. - A vignette on data upscaling which explains how to leverage
funbiogeoto aggregate automatically your data to coarser grain and use them in further analyses.
Citation
For the moment funbiogeo doesn’t offer a companion paper nor is it on CRAN. But if you happen to use it in your paper you can cite the package through:
Casajus N & Grenié M (2024). funbiogeo: Functional Biogeography Analyses. R package version 0.0.0.9000, https://github.com/frbcesab/funbiogeo.
You can also run:
citation("funbiogeo")Contributing
All types of contributions are encouraged and valued. For more information, check out our Contributor Guidelines.
Please note that the funbiogeo project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Acknowledgments
This package has been developed for the FRB-CESAB working group FREE and its followup FREE 2 that aims to advance the concept of functional rarity and examine the causes and consequences of functional rarity from local to global scales.
References
Monnet, AC., Cilleros, K., Médail, F. et al. WOODIV, a database of occurrences, functional traits, and phylogenetic data for all Euro-Mediterranean trees. Sci Data 8, 89 (2021). https://doi.org/10.1038/s41597-021-00873-3
Violle C, Reich, PB Pacala SW, et al. (2014) The emergence and promise of functional biogeography. Proceedings of the National Academy of Sciences, 111, 13690–13696. DOI: 10.1073/pnas.1415442111