The goal of the R package popbayes is to fit population
trajectories over time from counts of individuals collected at various
dates and with a variety of methods. It does so under a Bayesian
framework where the primary quantity being modeled is the rate of
increase between successive years (or any other time units for that
matter, the one used for date). The package can deal with multiple
species and multiple locations presented in a single data set, but each
count series made of the counts relative to one species
at one location will be processed independently.
The strength of popbayes is to handle, in a single
series, counts collected under different types of surveys (aerial vs
ground surveys), and estimated by different census methods (total
counts, sampling counts, and even guesstimates [i.e. expert
estimates]).
Before using this package, users need to install the freeware JAGS.
The workflow of popbayes consists in three main
steps:
- Formatting data (
format_data()) - Fitting trends (
fit_trend()) - Visualizing results (
plot_trend())
The package also provides a lot of functions to handle individual count series and model outputs. The following figure shows a more complete usage of the package.
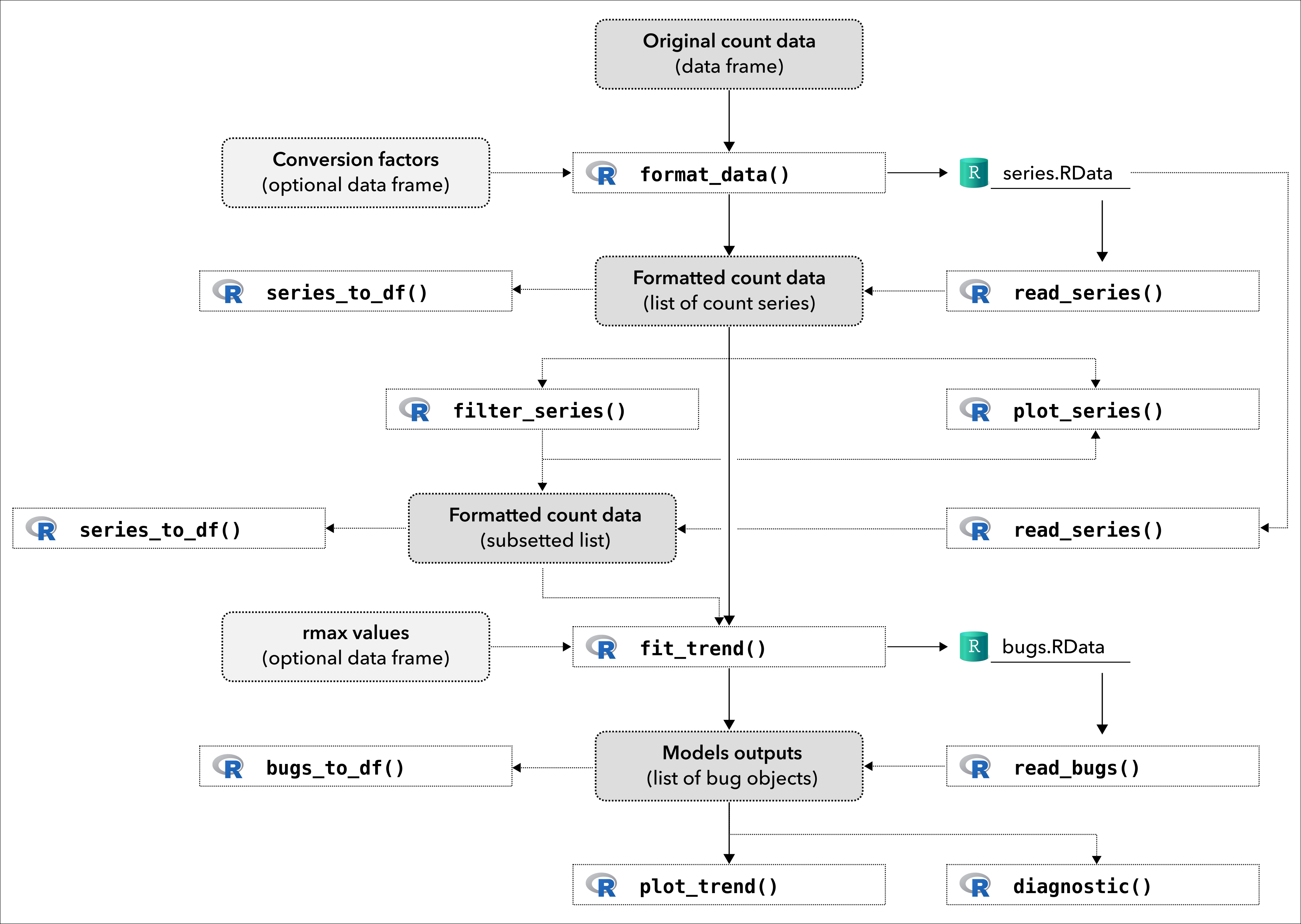
Framework of popbayes
The Garamba dataset
The package popbayes comes with an example dataset:
garamba. It contains counts of individuals from 10 African
mammal species surveyed in the Garamba National Park (Democratic
Republic of the Congo) from 1976 to 2017.
## Define filename path ----
file_path <- system.file("extdata", "garamba_survey.csv", package = "popbayes")
## Read CSV file ----
garamba <- read.csv(file = file_path)| location | species | date | stat_method | field_method | count | lower_ci | upper_ci | pref_field_method | conversion_A2G | rmax |
|---|---|---|---|---|---|---|---|---|---|---|
| Garamba | Alcelaphus buselaphus | 1976 | S | A | 7750 | 6280 | 9220 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1983 | S | A | 1932 | 1120 | 2744 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1984 | S | A | 1224 | 782 | 1666 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1986 | S | A | 1705 | 1116 | 2294 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1991 | S | A | 987 | 663 | 1311 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1993 | S | A | 3444 | 1290 | 5598 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1995 | S | A | 2819 | 1620 | 4018 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 1998 | S | A | 1685 | 1287 | 2083 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2000 | S | A | 1169 | 945 | 1393 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2002 | S | A | 1139 | 907 | 1371 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2003 | S | A | 1595 | 1142 | 2048 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2004 | S | A | 1204 | 811 | 1597 | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2012 | T | A | 552 | NA | NA | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2014 | T | A | 698 | NA | NA | G | 2.302 | 0.2748 |
| Garamba | Alcelaphus buselaphus | 2017 | T | A | 1051 | NA | NA | G | 2.302 | 0.2748 |
| Garamba | Giraffa camelopardalis | 1976 | S | A | 350 | 100 | 600 | A | 3.011 | 0.1750 |
| Garamba | Giraffa camelopardalis | 1983 | S | A | 175 | 12 | 338 | A | 3.011 | 0.1750 |
| Garamba | Giraffa camelopardalis | 1984 | S | A | 237 | 93 | 381 | A | 3.011 | 0.1750 |
| Garamba | Giraffa camelopardalis | 1986 | S | A | 153 | 13 | 293 | A | 3.011 | 0.1750 |
| Garamba | Giraffa camelopardalis | 1991 | S | A | 346 | 143 | 549 | A | 3.011 | 0.1750 |
This dataset has a typical structure with a location field
(location), a species name field (species), a
date field (date), and a count field
(count).
Statistical method
In addition to the fields location,
species, date, and count, a
fourth field is mandatory: stat_method.
This field specifies the census method that produced the count. It can
be T for a total count, X for a guesstimate
(i.e. expert estimate), or S for a sampling count.
To be usable by the Bayesian model, individual counts are to be accompanied by information on precision in the form of a 95% confidence interval. If counts are :
-
TorX, a confidence interval will be computed automatically by the functionformat_data()according respectively to the following formulas:
\[ CI_{(T)} = [\ 0.95 \times count\ ; 1.20 \times count\ ] \] \[ CI_{(X)} = [\ 0.80 \times count\ ; 1.20 \times count\ ] \]
-
S, users need to supply a measure of precision. Precision is preferably provided in the form of a 95% CI by means of two fields:lower_ciandupper_ci(as in thegarambadataset). Alternatively, it may also be given in the form of a standard deviation (sd), a variance (var), or a coefficient of variation (cv). Note that precision metrics can be different between counts. For instance, someScounts may have ansdvalue and otherslower_ciandupper_ci. In that case, three precision columns would be required (lower_ci,upper_ci, andsd). AnScount with no measure of precision will be detected as an anomaly byformat_data()by default. The optionna.rm = TRUEmay be used to automatically remove such counts from the series. If it is desirable to maintain such counts in the count series, we suggest to enter a value for the coefficient of variation, e.g. the average coefficient of variation of the other counts in the series.
Field method
Another optional column, field_method,
may be provided. It refers to the type of survey used to collect data.
This can be A for aerial survey or G for
ground survey. This column becomes mandatory as soon as both field
methods are present in a series.
The detectability of a species is indeed strongly dependent on the
survey method and each species has its own preferred field
method, the one that is assumed to provide estimates closer to the
truth. So, even if a series is homogeneous relative to the
field method, it is recommended to provide the column
field_method if counts have been collected under the not
preferred field method. That will force conversion towards the
preferred field method.
Count conversion
The function format_data() will convert counts (and 95%
CI bounds) into their equivalent in the preferred field method for the
species. To this aim, two pieces of information are required :
-
pref_field_method: the preferred field method for the species (AorG); -
conversion_A2G: the multiplicative factor used to convert an aerial count into an equivalent ground count.
The package popbayes provides the
species_info dataset, which contains these two pieces of
information for 15 African mammal species.
data("species_info")| species | category | pref_field_method | conversion_A2G | rmax |
|---|---|---|---|---|
| Aepyceros melampus | MLB | G | 6.747 | 0.4010 |
| Alcelaphus buselaphus | LLB | G | 2.302 | 0.2748 |
| Connochaetes taurinus | LLB | G | 2.302 | 0.2679 |
| Damaliscus lunatus | MLB | G | 6.747 | 0.2990 |
| Eudorcas rufifrons | MLB | G | 6.747 | 0.5270 |
| Giraffa camelopardalis | Giraffe | A | 3.011 | 0.1750 |
| Hippotragus equinus | LLB | G | 2.302 | 0.2420 |
| Kobus ellipsiprymnus | MLB | G | 6.747 | 0.2702 |
| Kobus kob | MLB | G | 6.747 | 0.3802 |
| Loxodonta africana | Elephant | A | 0.659 | 0.1120 |
| Ourebia ourebi | MLB | G | 6.747 | 0.5988 |
| Redunca redunca | MLB | G | 6.747 | 0.4010 |
| Syncerus caffer | LD | A | 0.561 | 0.2080 |
| Tragelaphus derbianus | LLB | G | 2.302 | 0.1500 |
| Tragelaphus scriptus | MLB | G | 6.747 | 0.4487 |
If users work only with species in this table, the package
popbayes can automatically retrieve the values of
pref_field_method and conversion_A2G from the
species_info data set. But for other species, users
need to supply the information themselves when running
format_data(). These values may be provided as additional
fields in the count data set. Care must then be taken that the same
value is consistently repeated for each count of the same species. For
users with sufficient command of R, we recommend rather to create an
independent additional table similar to species_info and to
pass it to the function format_data() as the data frame
argument info.
Note: Currently format_data() takes its
information for count conversion from one source only
with priority given to info, then to additional fields in
data (if info is not provided), and eventually to the
species_info table of the package (when the other two
sources are lacking). That means that the source with the highest
priority must be complete with respect to the species present in data,
as it will be used exclusively to any other source. If, say, you use
info, you cannot expect format_data() to
retrieve conversion information for a species undocumented in
info from the species_info table of the
package. However, you can easily construct info from a copy
of species_info, which additionally provides a ready
template. It suffices to add any species not already in
species_info as shown below.
Let’s assume that, in addition to other species present in the
package species_info table, we have counts of
Taurotragus oryx and Taurotragus derbianus. We can
construct info as follows.
## Extract the relevant columns of the package table "species_info" ----
info_from_package <- species_info[ , c("species", "pref_field_method", "conversion_A2G", "rmax")]
## Add the new species ----
new_conversion_info <- data.frame("species" = c("Taurotragus oryx","Taurotragus derbianus"),
"pref_field_method" = "G",
"conversion_A2G" = 2.302,
"rmax" = 0.1500)
## Append the new species ----
info <- rbind(info_from_package, new_conversion_info)
info
#> species pref_field_method conversion_A2G rmax
#> 1 Aepyceros melampus G 6.747 0.4010
#> 2 Alcelaphus buselaphus G 2.302 0.2748
#> 3 Connochaetes taurinus G 2.302 0.2679
#> 4 Damaliscus lunatus G 6.747 0.2990
#> 5 Eudorcas rufifrons G 6.747 0.5270
#> 6 Giraffa camelopardalis A 3.011 0.1750
#> 7 Hippotragus equinus G 2.302 0.2420
#> 8 Kobus ellipsiprymnus G 6.747 0.2702
#> 9 Kobus kob G 6.747 0.3802
#> 10 Loxodonta africana A 0.659 0.1120
#> 11 Ourebia ourebi G 6.747 0.5988
#> 12 Redunca redunca G 6.747 0.4010
#> 13 Syncerus caffer A 0.561 0.2080
#> 14 Tragelaphus derbianus G 2.302 0.1500
#> 15 Tragelaphus scriptus G 6.747 0.4487
#> 16 Taurotragus oryx G 2.302 0.1500
#> 17 Taurotragus derbianus G 2.302 0.1500If you do not have conversion information of your own for a new
species, you can rely on the conversion information of species with
similar characteristics (for example the two Taurotragus
species belong to the category LLB). The package popbayes
distinguishes five categories of species:
- MLB: Medium-sized Light and Brown species (20-150kg)
- LLB: Large Light and Brown species (>150kg)
- LD: Large Dark (>150kg)
- Elephant
- Giraffe
The field category of the species_info
table indicates which species belong to each.
Relative rate of increase
The demographic potential of a species is limited. The intrinsic rate
of increase (called rmax) is the maximum increase in log
population size that a species can attain in a year.
We strongly recommend using the rmax values while
estimating population trend to limit yearly population growth estimated
by the model (the default).
As for pref_field_method and
conversion_A2G, rmax values (specific to a
species) can be provided in an additional field of the count dataset
(garamba), as additional field of the info
data frame, or internally can be retrieved from the internal dataset of
popbayes.
How to find the species rmax value?
According to Sinclair (2003), rmax is related to the
body mass of adult females W by the formula:
\[ rmax = 1.375 \times W^{-0.315} \]
Body masses are found in the literature in publications such as Kingdon & Hoffman (2013), Cornelis et al. (2014), Illius & Gordon (1992), Sinclair (1996), Suraud et al. (2012), or Foley & Faust (2010).
If you know the body mass of adult females of the species, you can
compute the rmax value with the function
w_to_rmax().
Alternatively, rmax can be obtained from previous
demographic analyses.
Important note: The intrinsic rate of increase refers to a change over one year. If a different time unit is used for the dates (say a month), the rmax to provide must be adapted (here divided by 12). The rmax values in popbayes cannot be used for time units other than one year.
Checking data
The first thing that the function format_data() does is
to check the validity of the content of the different fields of the
count data set. Here we will explore our data to avoid errors when using
the function format_data().
In particular, we need to check location and
species spelling, date and count
field format, and the stat_method and
field_method categories.
Check location field
unique(garamba$"location")
#> [1] "Garamba"
sum(is.na(garamba$"location")) # Are there any missing values?
#> [1] 0Field location can be either a
character or a factor. It
cannot contain any NA values.
Check species field
unique(garamba$"species")
#> [1] "Alcelaphus buselaphus" "Giraffa camelopardalis" "Hippotragus equinus"
#> [4] "Kobus ellipsiprymnus" "Kobus kob" "Loxodonta africana"
#> [7] "Ourebia ourebi" "Redunca redunca" "Syncerus caffer"
#> [10] "Tragelaphus scriptus"
sum(is.na(garamba$"species")) # Are there any missing values?
#> [1] 0
## Are there species absent from the 'species_info' popbayes dataset?
garamba_species <- unique(garamba$"species")
garamba_species[which(!(garamba_species %in% species_info$"species"))]
#> character(0)Field species can be either a
character or a factor. It
cannot contain any NA values.
Check date field
is.numeric(garamba$"date") # Are dates in a numerical format?
#> [1] TRUE
sum(is.na(garamba$"date")) # Are there any missing values?
#> [1] 0
range(garamba$"date") # What is the temporal extent?
#> [1] 1976 2017Field date must be a
numeric. It cannot contain any
NA values. This said, the time unit is arbitrary, and
fractional values of years (or another unit) are allowed. As long as
numeric values are entered, the package will work.
On the other hand, if you have a date format (e.g. ‘2021/05/19’), you need to convert it to a numeric format. For instance:
## Convert a character to a date object ----
x <- as.Date("2021/05/19")
x
#> [1] "2021-05-19"
## Convert a date to a numeric (number of days since 1970/01/01) ----
x <- as.numeric(x)
x
#> [1] 18766
## Check ----
as.Date(x, origin = as.Date("1970/01/01"))
#> [1] "2021-05-19"Other methods exist to convert a date to a
numeric format. You may prefer computing the number of days
since the first date of your survey. It’s up to you.
Check count field
is.numeric(garamba$"count") # Are counts in a numerical format?
#> [1] TRUE
range(garamba$"count") # What is the range of values?
#> [1] 0 53312
sum(is.na(garamba$"count")) # Are there any missing values?
#> [1] 0Field count must be a
positive numeric (zero counts are
allowed). NA counts cannot be used for fitting trends. The
format_data() function (see below) has an option for
dropping them.
Check stat_method field
unique(garamba$"stat_method")
#> [1] "S" "T"
sum(is.na(garamba$"stat_method")) # Are there any missing values?
#> [1] 0Field stat_method can be either a
character or a factor. It
must contain only T, X, or
S categories and cannot contain any
NA values.
Check field_method field
unique(garamba$"field_method")
#> [1] "A"
sum(is.na(garamba$"field_method")) # Are there any missing values?
#> [1] 0Field field_method can be either a
character or a factor. It
must contain only A, or G
categories and cannot contain any NA
values.
Formatting data
This first popbayes function to use is
format_data(). This function provides an easy way to get
individual count series ready to be analyzed by the package. It must be
used prior to all other functions.
First let’s define the path (relative or absolute) to save objects/results, namely the formatted count series that can be extracted from the data set.
path <- "the_folder_to_store_outputs"The function format_data() has many arguments to provide
the names of the columns in the user’s dataset that contain
location, species, lower_ci, etc.
By default column names are the same as in the Garamba dataset. If your
location field, say, is “site”, you’ll need to use the argument
location as follows: location = "site".
garamba_formatted <- popbayes::format_data(data = garamba,
path = path,
field_method = "field_method",
pref_field_method = "pref_field_method",
conversion_A2G = "conversion_A2G",
rmax = "rmax")
#> ✔ Detecting 10 count series.As said above, if you have to add your own count conversion data, you
need specify the names of columns for the preferred field method, the
conversion factor, and rmax as this:
pref_field_method = "column_with_preferred_field_method",
conversion_A2G = "column_with_conversion_A2Gor",
rmax = "column_with_conversion_rmax", or alternatively use
the argument info:
info = "dataframe_with_conversion_info".
Let’s explore the output.
## Class of the object ----
class(garamba_formatted)
#> [1] "list"
## Number of elements (i.e. number of count series) ----
length(garamba_formatted)
#> [1] 10
## Get series names ----
popbayes::list_series(path)
#> [1] "garamba__alcelaphus_buselaphus" "garamba__giraffa_camelopardalis"
#> [3] "garamba__hippotragus_equinus" "garamba__kobus_ellipsiprymnus"
#> [5] "garamba__kobus_kob" "garamba__loxodonta_africana"
#> [7] "garamba__ourebia_ourebi" "garamba__redunca_redunca"
#> [9] "garamba__syncerus_caffer" "garamba__tragelaphus_scriptus"Let’s work with the count series
"garamba__alcelaphus_buselaphus". We can use the function
filter_series().
## Retrieve series by species and location ----
a_buselaphus <- popbayes::filter_series(data = garamba_formatted,
species = "Alcelaphus buselaphus",
location = "Garamba")
#> ✔ Found 1 series with "Alcelaphus buselaphus" and "Garamba".Let’s display the series content.
print(a_buselaphus)
#> $garamba__alcelaphus_buselaphus
#> $garamba__alcelaphus_buselaphus$location
#> [1] "Garamba"
#>
#> $garamba__alcelaphus_buselaphus$species
#> [1] "Alcelaphus buselaphus"
#>
#> $garamba__alcelaphus_buselaphus$dates
#> [1] 1976 1983 1984 1986 1991 1993 1995 1998 2000 2002 2003 2004 2012 2014 2017
#>
#> $garamba__alcelaphus_buselaphus$n_dates
#> [1] 15
#>
#> $garamba__alcelaphus_buselaphus$stat_methods
#> [1] "S" "T"
#>
#> $garamba__alcelaphus_buselaphus$field_methods
#> [1] "A"
#>
#> $garamba__alcelaphus_buselaphus$pref_field_method
#> [1] "G"
#>
#> $garamba__alcelaphus_buselaphus$conversion_A2G
#> [1] 2.302
#>
#> $garamba__alcelaphus_buselaphus$rmax
#> [1] 0.2748
#>
#> $garamba__alcelaphus_buselaphus$data_original
#> location species date stat_method field_method
#> 1 Garamba Alcelaphus buselaphus 1976 S A
#> 2 Garamba Alcelaphus buselaphus 1983 S A
#> 3 Garamba Alcelaphus buselaphus 1984 S A
#> 4 Garamba Alcelaphus buselaphus 1986 S A
#> 5 Garamba Alcelaphus buselaphus 1991 S A
#> 6 Garamba Alcelaphus buselaphus 1993 S A
#> 7 Garamba Alcelaphus buselaphus 1995 S A
#> 8 Garamba Alcelaphus buselaphus 1998 S A
#> 9 Garamba Alcelaphus buselaphus 2000 S A
#> 10 Garamba Alcelaphus buselaphus 2002 S A
#> 11 Garamba Alcelaphus buselaphus 2003 S A
#> 12 Garamba Alcelaphus buselaphus 2004 S A
#> 13 Garamba Alcelaphus buselaphus 2012 T A
#> 14 Garamba Alcelaphus buselaphus 2014 T A
#> 15 Garamba Alcelaphus buselaphus 2017 T A
#> pref_field_method conversion_A2G rmax count_orig lower_ci_orig
#> 1 G 2.302 0.2748 7750 6280
#> 2 G 2.302 0.2748 1932 1120
#> 3 G 2.302 0.2748 1224 782
#> 4 G 2.302 0.2748 1705 1116
#> 5 G 2.302 0.2748 987 663
#> 6 G 2.302 0.2748 3444 1290
#> 7 G 2.302 0.2748 2819 1620
#> 8 G 2.302 0.2748 1685 1287
#> 9 G 2.302 0.2748 1169 945
#> 10 G 2.302 0.2748 1139 907
#> 11 G 2.302 0.2748 1595 1142
#> 12 G 2.302 0.2748 1204 811
#> 13 G 2.302 0.2748 552 NA
#> 14 G 2.302 0.2748 698 NA
#> 15 G 2.302 0.2748 1051 NA
#> upper_ci_orig
#> 1 9220
#> 2 2744
#> 3 1666
#> 4 2294
#> 5 1311
#> 6 5598
#> 7 4018
#> 8 2083
#> 9 1393
#> 10 1371
#> 11 2048
#> 12 1597
#> 13 NA
#> 14 NA
#> 15 NA
#>
#> $garamba__alcelaphus_buselaphus$data_converted
#> location species date stat_method field_method
#> 1 Garamba Alcelaphus buselaphus 1976 S A
#> 2 Garamba Alcelaphus buselaphus 1983 S A
#> 3 Garamba Alcelaphus buselaphus 1984 S A
#> 4 Garamba Alcelaphus buselaphus 1986 S A
#> 5 Garamba Alcelaphus buselaphus 1991 S A
#> 6 Garamba Alcelaphus buselaphus 1993 S A
#> 7 Garamba Alcelaphus buselaphus 1995 S A
#> 8 Garamba Alcelaphus buselaphus 1998 S A
#> 9 Garamba Alcelaphus buselaphus 2000 S A
#> 10 Garamba Alcelaphus buselaphus 2002 S A
#> 11 Garamba Alcelaphus buselaphus 2003 S A
#> 12 Garamba Alcelaphus buselaphus 2004 S A
#> 13 Garamba Alcelaphus buselaphus 2012 T A
#> 14 Garamba Alcelaphus buselaphus 2014 T A
#> 15 Garamba Alcelaphus buselaphus 2017 T A
#> pref_field_method conversion_A2G rmax count_conv lower_ci_conv
#> 1 G 2.302 0.2748 17840.500 14456.560
#> 2 G 2.302 0.2748 4447.464 2578.240
#> 3 G 2.302 0.2748 2817.648 1800.164
#> 4 G 2.302 0.2748 3924.910 2569.032
#> 5 G 2.302 0.2748 2272.074 1526.226
#> 6 G 2.302 0.2748 7928.088 2969.580
#> 7 G 2.302 0.2748 6489.338 3729.240
#> 8 G 2.302 0.2748 3878.870 2962.674
#> 9 G 2.302 0.2748 2691.038 2175.390
#> 10 G 2.302 0.2748 2621.978 2087.914
#> 11 G 2.302 0.2748 3671.690 2628.884
#> 12 G 2.302 0.2748 2771.608 1866.922
#> 13 G 2.302 0.2748 1270.704 1207.169
#> 14 G 2.302 0.2748 1606.796 1526.456
#> 15 G 2.302 0.2748 2419.402 2298.432
#> upper_ci_conv field_method_conv
#> 1 21224.440 G
#> 2 6316.688 G
#> 3 3835.132 G
#> 4 5280.788 G
#> 5 3017.922 G
#> 6 12886.596 G
#> 7 9249.436 G
#> 8 4795.066 G
#> 9 3206.686 G
#> 10 3156.042 G
#> 11 4714.496 G
#> 12 3676.294 G
#> 13 1524.845 G
#> 14 1928.155 G
#> 15 2903.282 GThe first elements of the list provide a summary of the count series.
If we compare the two last data frames (data_original
and data_converted), they are not identical. The function
format_data() has 1) computed 95% CI
boundaries for total counts (coded T in the column
stat_method), and 2) converted all counts
(and CI boundaries) to their equivalent in the preferred field method
(from A to G) by applying the conversion
factor of 2.302.
The Bayesian model will use counts and precision measures from the
data_converted data frame.
Before fitting the population size trend we can visualize the count
series with plot_series().
popbayes::plot_series("garamba__alcelaphus_buselaphus", path = path)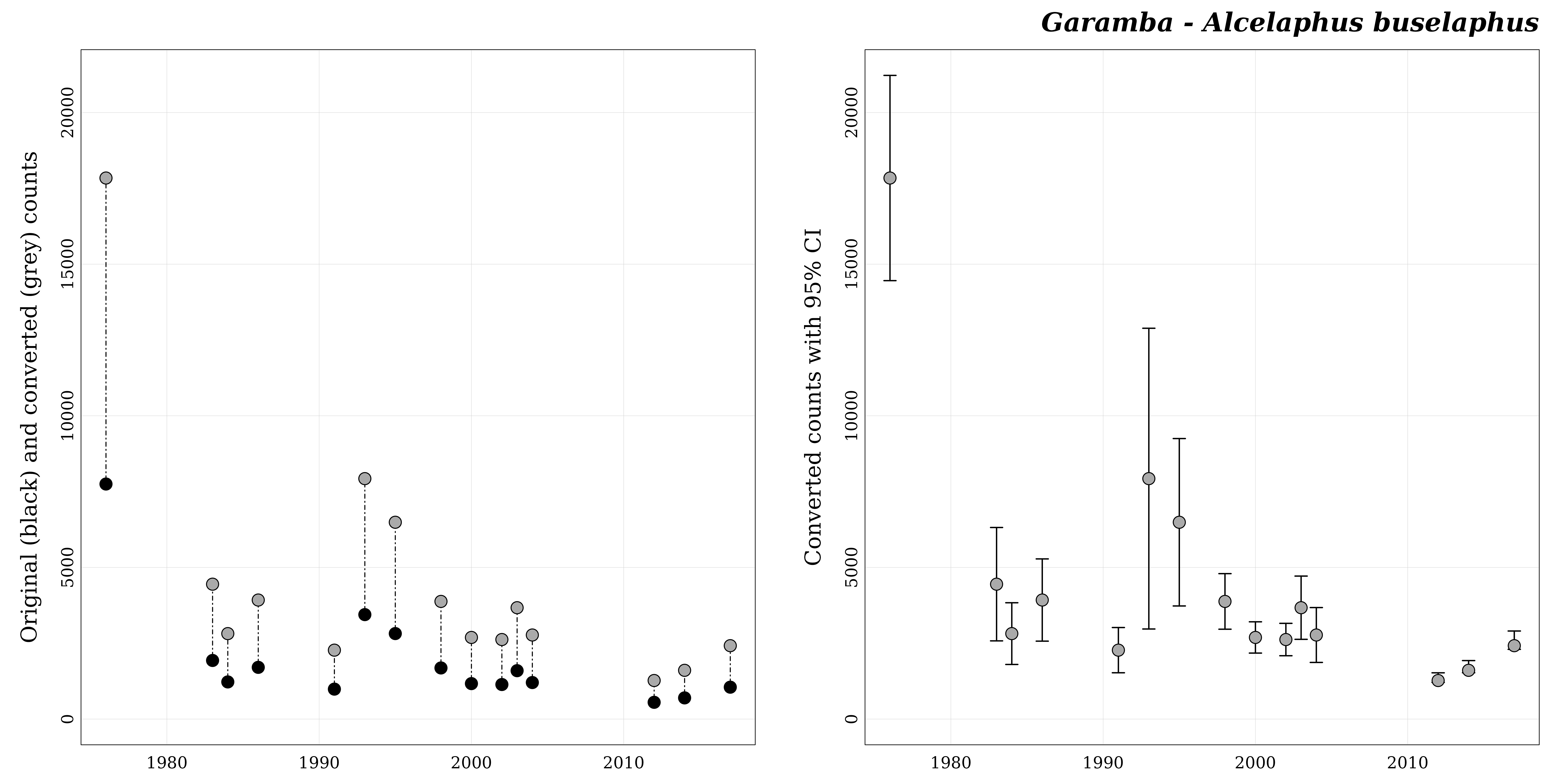
The function format_data() has also exported the count
series as .RData files in the path folder
where they have been dispatched into sub-folders, one per series.
list.files(path, recursive = TRUE)#> [1] "garamba__alcelaphus_buselaphus/garamba__alcelaphus_buselaphus_data.RData"
#> [2] "garamba__giraffa_camelopardalis/garamba__giraffa_camelopardalis_data.RData"
#> [3] "garamba__hippotragus_equinus/garamba__hippotragus_equinus_data.RData"
#> [4] "garamba__kobus_ellipsiprymnus/garamba__kobus_ellipsiprymnus_data.RData"
#> [5] "garamba__kobus_kob/garamba__kobus_kob_data.RData"
#> [6] "garamba__loxodonta_africana/garamba__loxodonta_africana_data.RData"
#> [7] "garamba__ourebia_ourebi/garamba__ourebia_ourebi_data.RData"
#> [8] "garamba__redunca_redunca/garamba__redunca_redunca_data.RData"
#> [9] "garamba__syncerus_caffer/garamba__syncerus_caffer_data.RData"
#> [10] "garamba__tragelaphus_scriptus/garamba__tragelaphus_scriptus_data.RData"These *_data.RData files (count series) can be imported
later by running the function read_series().
a_buselaphus <- popbayes::read_series("garamba__alcelaphus_buselaphus", path = path)Fitting trend
The function fit_trend() fits population trajectories
over time from counts of individuals formatted by
format_data(). It does so under a Bayesian framework where
the primary quantity being modeled is the annual rate of increase (more
generally, the rate of increase per the time unit used for dates).
This function only works on the output of format_data()
(or filter_series()).
Here is the default usage of the function
fit_trend():
a_buselaphus_bugs <- popbayes::fit_trend(a_buselaphus, path = path)The function returns an n-element list (where n is the number of
count series). Each element of the list is a BUGS output as provided by
JAGS. It has also exported these BUGS outputs as .RData
files in the path folder where they have been dispatched
into sub-folders, one per series.
These *_bugs.RData files (BUGS outputs) can be imported
later by running the function read_bugs().
a_buselaphus_bugs <- popbayes::read_bugs("garamba__alcelaphus_buselaphus", path = path)The function diagnostic() allows to check if estimation
of all parameters of the model has converged. This diagnostic is
performed by comparing the Rhat value of each parameter to
a threshold (default is 1.1).
popbayes::diagnostic(a_buselaphus_bugs)
#> All models have converged.In case convergence was not reached for some series, we suggest
rerunning fit_trend() on these series after increasing the
number of iterations (ni) and possibly the number of
initial iterations discarded (nb) from their respective
defaults of 50,000 and 10,000. For example:
a_buselaphus_bugs <- popbayes::fit_trend(a_buselaphus, path = path, ni = 100000, nb = 20000)This process may be repeated with increasing values of ni and nb until convergence is eventually reached.
Finally we can use the function plot_trend() to
visualize model predictions and estimated yearly relative growth
rates.
popbayes::plot_trend("garamba__alcelaphus_buselaphus", path = path)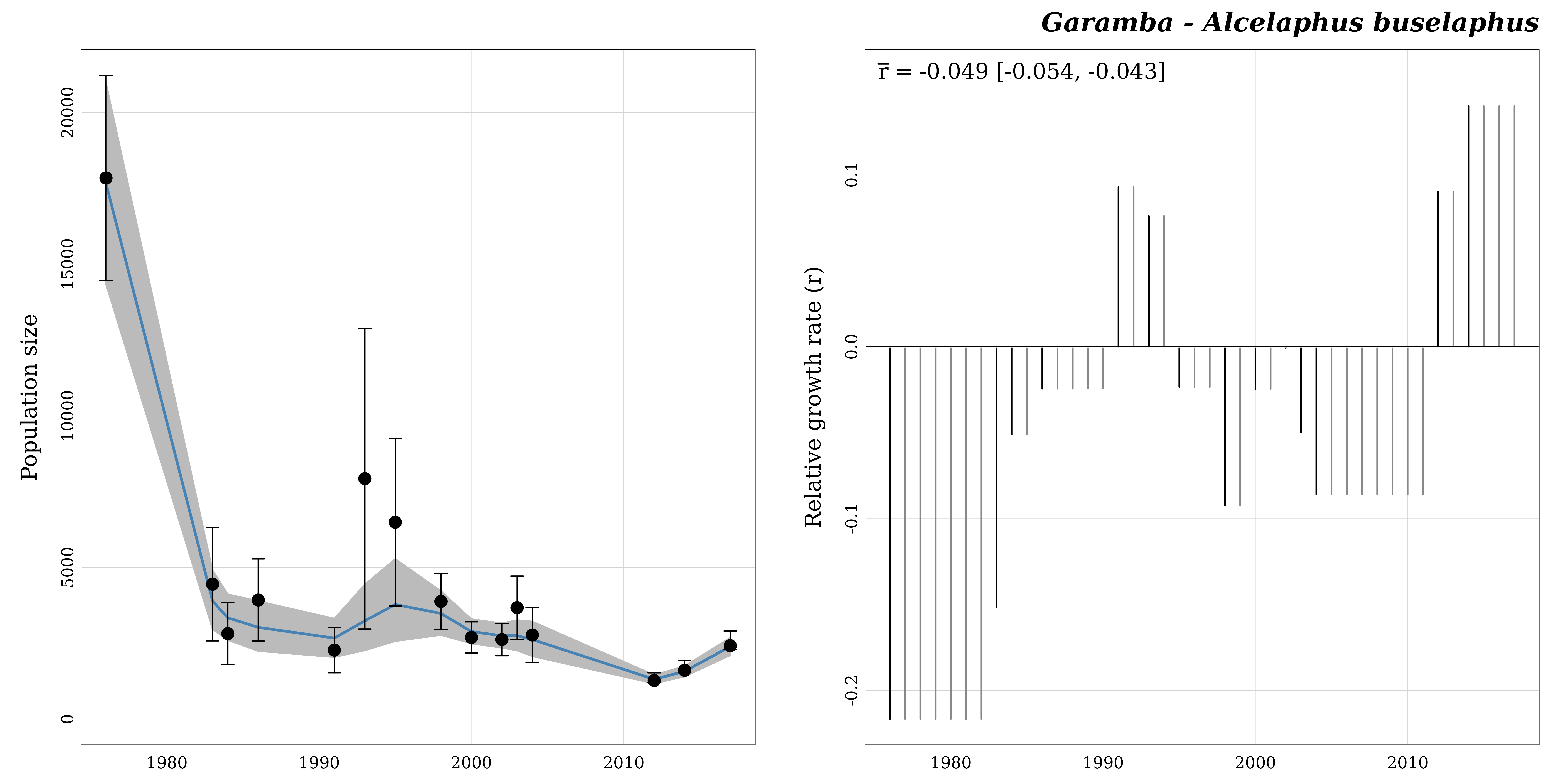
References
Cornelis D et al. (2014) Species account: African buffalo (Syncerus caffer). In: Ecology, Evolution and Behaviour of Wild Cattle: Implications for Conservation (Eds M Melletti & J Burton). Cambridge University Press, Cambridge. DOI: 10.1017/CBO9781139568098.
Foley CAH & Faust LJ (2010) Rapid population growth in an elephant Loxodonta africana population recovering from poaching in Tarangire National Park, Tanzania. Oryx, 44, 205-212. DOI: 10.1017/S0030605309990706.
Illius AW & Gordon IJ (1992) Modelling the nutritional ecology of ungulate herbivores: evolution of body size and competitive interactions. Oecologia, 89, 428-434. DOI: 10.1017/S0030605309990706.
Kingdon J & Hoffman M (2013) Mammals of Africa. Volume VI: Pigs, Hippopotamuses, Chevrotain, Giraffes, Deer and Bovids. Bloomsbury Publishing, London, United Kingdom, 680 pp.
Sinclair ARE (1996) Mammal populations: fluctuation, regulation, life history theory, and their implications for conservation. In: Frontiers of population ecology (Eds RB Floyd & AW Sheppard), pp. 127-154. CSIRO: Melbourne, Australia.
Sinclair ARE (2003) Mammal population regulation, keystone processes and ecosystem dynamics. Philosophical Transactions: Biological Sciences, 358, 1729-1740. DOI: 10.1098/rstb.2003.1359.
Suraud JP et al. (2012) Higher than expected growth rate of the endangered West African giraffe Giraffa camelopardalis peralta: a successful human-wildlife cohabitation. Oryx, 46, 577-583. DOI: 10.1017/S0030605311000639.
